The Science
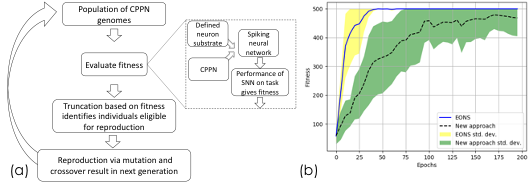
Researchers from Oak Ridge National Laboratory and the University of Central Florida have extended an evolutionary approach for training spiking neural networks (SNNs) by implementing an indirect encoding of SNN individuals in the population. Specifically, they evolved SNNs using Compositional Pattern Producing Networks (CPPNs) to learn connectivity patterns between neurons defined in coordinate spaces.
Utilizing CPPNs rather than direct encodings to evolve parameters allows small genome representations (on the order of 10s to 100s of genes) to potentially represent thousands to millions of SNN parameter values.
The Impact
With indirect encodings, significantly larger networks with more complex connectivity can be evolved, enabling broader applicability to more complex applications, such as classifying large scientific images.
PI/Facility Lead(s): Catherine Schuman (ORNL CSMD)
ASCR Program/Facility: Early Career Program
Funding: ASCR
Publication(s) for this work: Elbrecht, Daniel and Catherine Schuman, "Neuroevolution of Spiking Neural Networks Using Compositional Pattern Producing Networks." International Conference on Neuromorphic Systems, July 28-30, 2020. DOI: 10.1145/3407197.3407198.


