Filter News
Area of Research
News Type
News Topics
- (-) Biology (5)
- Artificial Intelligence (7)
- Big Data (3)
- Bioenergy (2)
- Biomedical (4)
- Buildings (3)
- Chemical Sciences (2)
- Climate Change (4)
- Computer Science (10)
- Coronavirus (3)
- Critical Materials (1)
- Cybersecurity (1)
- Decarbonization (2)
- Energy Storage (3)
- Environment (3)
- Exascale Computing (6)
- Frontier (7)
- Grid (2)
- High-Performance Computing (8)
- Machine Learning (5)
- Materials (8)
- Materials Science (5)
- Microscopy (2)
- Nanotechnology (3)
- National Security (3)
- Neutron Science (1)
- Partnerships (1)
- Physics (1)
- Quantum Computing (7)
- Quantum Science (4)
- Security (2)
- Simulation (5)
- Space Exploration (1)
- Summit (7)
- Sustainable Energy (2)
Media Contacts
A new paper published in Nature Communications adds further evidence to the bradykinin storm theory of COVID-19’s viral pathogenesis — a theory that was posited two years ago by a team of researchers at the Department of Energy’s Oak Ridge National Laboratory.

ORNL scientists will present new technologies available for licensing during the annual Technology Innovation Showcase. The event is 9 a.m. to 3 p.m. Thursday, June 16, at the Manufacturing Demonstration Facility at ORNL’s Hardin Valley campus.

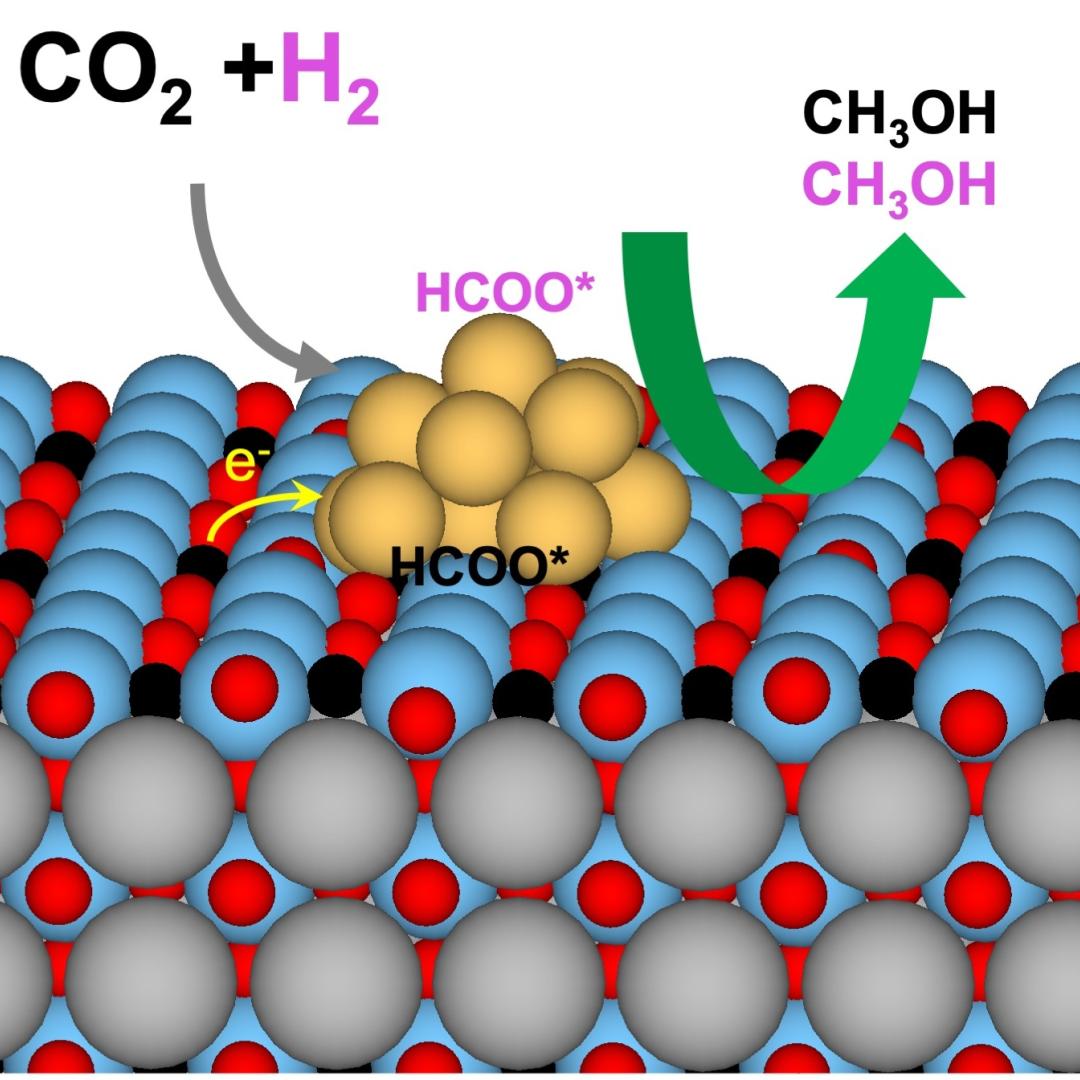
Tackling the climate crisis and achieving an equitable clean energy future are among the biggest challenges of our time.
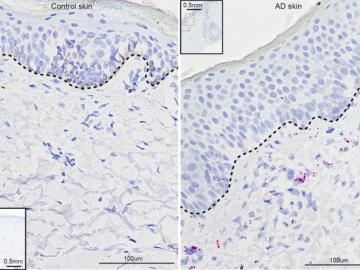
University of Pennsylvania researchers called on computational systems biology expertise at Oak Ridge National Laboratory to analyze large datasets of single-cell RNA sequencing from skin samples afflicted with atopic dermatitis.
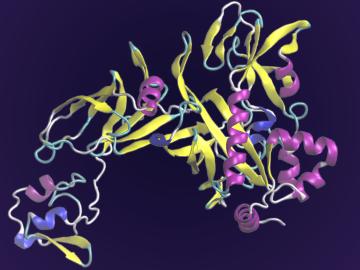
A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory and the Georgia Institute of Technology is using supercomputing and revolutionary deep learning tools to predict the structures and roles of thousands of proteins with unknown functions.