Filter News
Area of Research
News Topics
- (-) Coronavirus (2)
- (-) Decarbonization (3)
- 3-D Printing/Advanced Manufacturing (7)
- Advanced Reactors (1)
- Artificial Intelligence (1)
- Big Data (1)
- Bioenergy (10)
- Biology (14)
- Biomedical (4)
- Biotechnology (2)
- Buildings (1)
- Chemical Sciences (4)
- Clean Water (4)
- Climate Change (9)
- Composites (4)
- Computer Science (4)
- Critical Materials (5)
- Energy Storage (7)
- Environment (18)
- Fusion (2)
- Grid (2)
- High-Performance Computing (3)
- Hydropower (3)
- Isotopes (2)
- Machine Learning (1)
- Materials (13)
- Materials Science (19)
- Mercury (1)
- Microscopy (6)
- Molten Salt (1)
- Nanotechnology (8)
- Neutron Science (4)
- Nuclear Energy (3)
- Physics (2)
- Polymers (6)
- Quantum Computing (1)
- Quantum Science (1)
- Simulation (1)
- Space Exploration (1)
- Sustainable Energy (12)
- Transportation (7)
Media Contacts
Oak Ridge National Laboratory scientists led the development of a supply chain model revealing the optimal places to site farms, biorefineries, pipelines and other infrastructure for sustainable aviation fuel production.
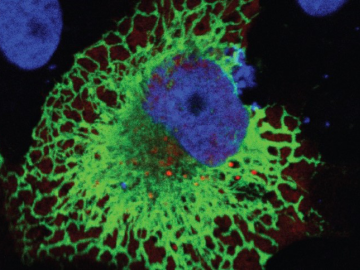
Oak Ridge National Laboratory scientists exploring bioenergy plant genetics have made a surprising discovery: a protein domain that could lead to new COVID-19 treatments.

ORNL researchers have identified specific proteins and amino acids that could control bioenergy plants’ ability to identify beneficial microbes that can enhance plant growth and storage of carbon in soils.

Oak Ridge National Laboratory scientists designed a recyclable polymer for carbon-fiber composites to enable circular manufacturing of parts that boost energy efficiency in automotive, wind power and aerospace applications.
Researchers from ORNL, the University of Tennessee at Chattanooga and Tuskegee University used mathematics to predict which areas of the SARS-CoV-2 spike protein are most likely to mutate.




