
Filter News
Area of Research
- (-) Biology and Environment (38)
- Advanced Manufacturing (2)
- Clean Energy (32)
- Computational Biology (2)
- Computational Engineering (3)
- Computer Science (5)
- Electricity and Smart Grid (1)
- Functional Materials for Energy (1)
- Fusion and Fission (23)
- Fusion Energy (13)
- Materials (23)
- Materials for Computing (4)
- Mathematics (1)
- National Security (18)
- Neutron Science (17)
- Nuclear Science and Technology (13)
- Supercomputing (76)
News Topics
- (-) Clean Water (11)
- (-) Coronavirus (13)
- (-) Exascale Computing (4)
- (-) Fusion (1)
- (-) Machine Learning (8)
- (-) Molten Salt (1)
- (-) Summit (10)
- 3-D Printing/Advanced Manufacturing (11)
- Advanced Reactors (1)
- Artificial Intelligence (9)
- Big Data (9)
- Bioenergy (45)
- Biology (73)
- Biomedical (16)
- Biotechnology (13)
- Buildings (2)
- Chemical Sciences (11)
- Climate Change (40)
- Composites (5)
- Computer Science (19)
- Critical Materials (1)
- Cybersecurity (1)
- Decarbonization (19)
- Energy Storage (7)
- Environment (89)
- Frontier (3)
- Grid (3)
- High-Performance Computing (20)
- Hydropower (8)
- Isotopes (2)
- Materials (12)
- Materials Science (6)
- Mathematics (3)
- Mercury (7)
- Microscopy (10)
- Nanotechnology (7)
- National Security (3)
- Net Zero (2)
- Neutron Science (4)
- Nuclear Energy (1)
- Partnerships (5)
- Physics (2)
- Polymers (2)
- Renewable Energy (1)
- Security (2)
- Simulation (14)
- Sustainable Energy (30)
- Transformational Challenge Reactor (1)
- Transportation (3)
Media Contacts
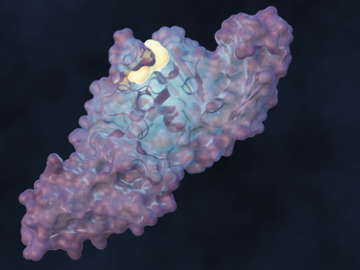
A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory designed a molecule that disrupts the infection mechanism of the SARS-CoV-2 coronavirus and could be used to develop new treatments for COVID-19 and other viral diseases.
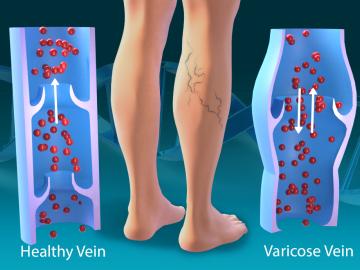
As part of a multi-institutional research project, scientists at ORNL leveraged their computational systems biology expertise and the largest, most diverse set of health data to date to explore the genetic basis of varicose veins.
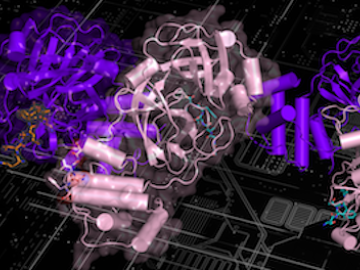
A new paper published in Nature Communications adds further evidence to the bradykinin storm theory of COVID-19’s viral pathogenesis — a theory that was posited two years ago by a team of researchers at the Department of Energy’s Oak Ridge National Laboratory.

Five technologies invented by scientists at the Department of Energy’s Oak Ridge National Laboratory have been selected for targeted investment through ORNL’s Technology Innovation Program.
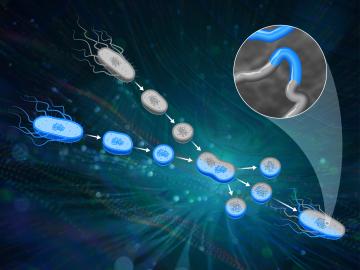
ORNL scientists had a problem mapping the genomes of bacteria to better understand the origins of their physical traits and improve their function for bioenergy production.

Surrounded by the mountains of landlocked Tennessee, Oak Ridge National Laboratory’s Teri O’Meara is focused on understanding the future of the vitally important ecosystems lining the nation’s coasts.

Spanning no less than three disciplines, Marie Kurz’s title — hydrogeochemist — already gives you a sense of the collaborative, interdisciplinary nature of her research at ORNL.
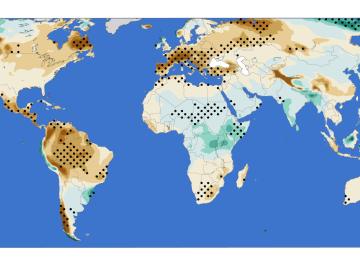
A new analysis from Oak Ridge National Laboratory shows that intensified aridity, or drier atmospheric conditions, is caused by human-driven increases in greenhouse gas emissions. The findings point to an opportunity to address and potentially reverse the trend by reducing emissions.
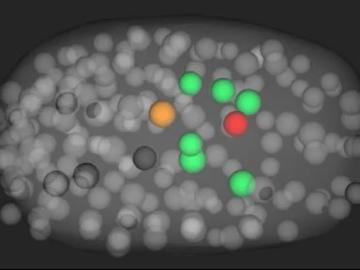
Scientists have developed a novel approach to computationally infer previously undetected behaviors within complex biological environments by analyzing live, time-lapsed images that show the positioning of embryonic cells in C. elegans, or roundworms. Their published methods could be used to reveal hidden biological activity.
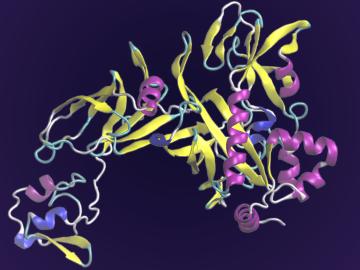
A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory and the Georgia Institute of Technology is using supercomputing and revolutionary deep learning tools to predict the structures and roles of thousands of proteins with unknown functions.


