Filter News
Area of Research
- (-) Biology and Environment (24)
- Advanced Manufacturing (1)
- Building Technologies (1)
- Clean Energy (26)
- Climate and Environmental Systems (1)
- Computational Biology (2)
- Computational Engineering (3)
- Computer Science (15)
- Fusion and Fission (2)
- Fusion Energy (2)
- Isotopes (1)
- Materials (17)
- Materials for Computing (8)
- Mathematics (1)
- National Security (20)
- Neutron Science (16)
- Nuclear Science and Technology (2)
- Quantum information Science (6)
- Supercomputing (112)
News Topics
- (-) Computer Science (19)
- (-) Summit (10)
- 3-D Printing/Advanced Manufacturing (11)
- Advanced Reactors (1)
- Artificial Intelligence (9)
- Big Data (9)
- Bioenergy (45)
- Biology (73)
- Biomedical (16)
- Biotechnology (13)
- Buildings (2)
- Chemical Sciences (11)
- Clean Water (11)
- Climate Change (40)
- Composites (5)
- Coronavirus (13)
- Critical Materials (1)
- Cybersecurity (1)
- Decarbonization (19)
- Energy Storage (7)
- Environment (89)
- Exascale Computing (4)
- Frontier (3)
- Fusion (1)
- Grid (3)
- High-Performance Computing (20)
- Hydropower (8)
- Isotopes (2)
- Machine Learning (8)
- Materials (12)
- Materials Science (6)
- Mathematics (3)
- Mercury (7)
- Microscopy (10)
- Molten Salt (1)
- Nanotechnology (7)
- National Security (3)
- Net Zero (2)
- Neutron Science (4)
- Nuclear Energy (1)
- Partnerships (5)
- Physics (2)
- Polymers (2)
- Renewable Energy (1)
- Security (2)
- Simulation (14)
- Sustainable Energy (30)
- Transformational Challenge Reactor (1)
- Transportation (3)
Media Contacts
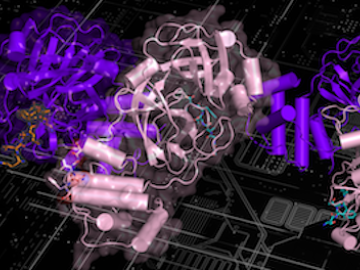
A new paper published in Nature Communications adds further evidence to the bradykinin storm theory of COVID-19’s viral pathogenesis — a theory that was posited two years ago by a team of researchers at the Department of Energy’s Oak Ridge National Laboratory.
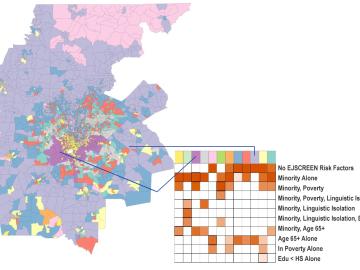
Scientists develop environmental justice lens to identify neighborhoods vulnerable to climate change
A new capability to identify urban neighborhoods, down to the block and building level, that are most vulnerable to climate change could help ensure that mitigation and resilience programs reach the people who need them the most.

Tackling the climate crisis and achieving an equitable clean energy future are among the biggest challenges of our time.
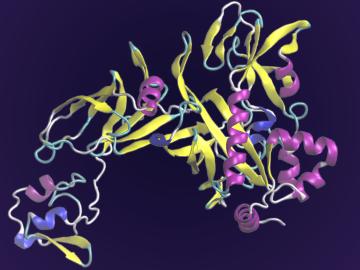
A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory and the Georgia Institute of Technology is using supercomputing and revolutionary deep learning tools to predict the structures and roles of thousands of proteins with unknown functions.

Improved data, models and analyses from ORNL scientists and many other researchers in the latest global climate assessment report provide new levels of certainty about what the future holds for the planet

RamSat’s mission is to take pictures of the forests around Gatlinburg, which were destroyed by wildfire in 2016. The mission is wholly designed and carried out by students, teachers and mentors, with support from numerous organizations, including Oak Ridge National Laboratory.

The Accelerating Therapeutics for Opportunities in Medicine , or ATOM, consortium today announced the U.S. Department of Energy’s Oak Ridge, Argonne and Brookhaven national laboratories are joining the consortium to further develop ATOM’s artificial intelligence, or AI-driven, drug discovery platform.
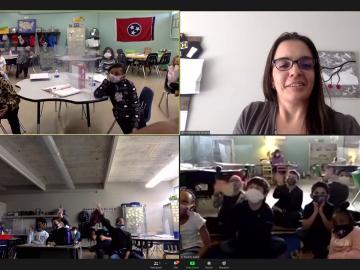
Twenty-seven ORNL researchers Zoomed into 11 middle schools across Tennessee during the annual Engineers Week in February. East Tennessee schools throughout Oak Ridge and Roane, Sevier, Blount and Loudon counties participated, with three West Tennessee schools joining in.

Six scientists at the Department of Energy’s Oak Ridge National Laboratory were named Battelle Distinguished Inventors, in recognition of obtaining 14 or more patents during their careers at the lab.
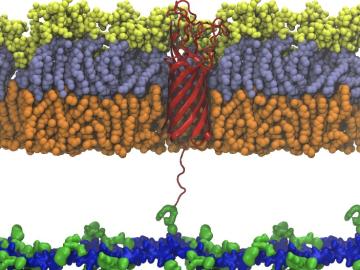
Scientists from Oak Ridge National Laboratory used high-performance computing to create protein models that helped reveal how the outer membrane is tethered to the cell membrane in certain bacteria.




