Filter News
Area of Research
- (-) Biology and Environment (16)
- (-) Supercomputing (17)
- Biological Systems (1)
- Clean Energy (6)
- Computational Biology (2)
- Computational Engineering (1)
- Fusion and Fission (1)
- Isotopes (5)
- Materials (7)
- Materials for Computing (2)
- National Security (2)
- Neutron Science (11)
- Nuclear Science and Technology (2)
News Topics
- (-) Biomedical (28)
- 3-D Printing/Advanced Manufacturing (13)
- Advanced Reactors (2)
- Artificial Intelligence (40)
- Big Data (25)
- Bioenergy (49)
- Biology (75)
- Biotechnology (14)
- Buildings (6)
- Chemical Sciences (14)
- Clean Water (11)
- Climate Change (51)
- Composites (5)
- Computer Science (104)
- Coronavirus (22)
- Critical Materials (4)
- Cybersecurity (9)
- Decarbonization (22)
- Energy Storage (11)
- Environment (102)
- Exascale Computing (24)
- Frontier (28)
- Fusion (2)
- Grid (7)
- High-Performance Computing (51)
- Hydropower (8)
- Isotopes (2)
- Machine Learning (18)
- Materials (24)
- Materials Science (22)
- Mathematics (3)
- Mercury (7)
- Microscopy (16)
- Molten Salt (1)
- Nanotechnology (16)
- National Security (9)
- Net Zero (3)
- Neutron Science (16)
- Nuclear Energy (5)
- Partnerships (5)
- Physics (8)
- Polymers (4)
- Quantum Computing (19)
- Quantum Science (24)
- Renewable Energy (1)
- Security (5)
- Simulation (23)
- Software (1)
- Space Exploration (3)
- Summit (46)
- Sustainable Energy (35)
- Transformational Challenge Reactor (1)
- Transportation (8)
Media Contacts

Scientists at ORNL used their expertise in quantum biology, artificial intelligence and bioengineering to improve how CRISPR Cas9 genome editing tools work on organisms like microbes that can be modified to produce renewable fuels and chemicals.

Hilda Klasky, an R&D staff member in the Scalable Biomedical Modeling group at ORNL, has been selected as a senior member of the Association of Computing Machinery, or ACM.

Scientist-inventors from ORNL will present seven new technologies during the Technology Innovation Showcase on Friday, July 14, from 8 a.m.–4 p.m. at the Joint Institute for Computational Sciences on ORNL’s campus.

Scientists at ORNL have confirmed that bacteria-killing viruses called bacteriophages deploy a sneaky tactic when targeting their hosts: They use a standard genetic code when invading bacteria, then switch to an alternate code at later stages of

Tomás Rush began studying the mysteries of fungi in fifth grade and spent his college intern days tromping through forests, swamps and agricultural lands searching for signs of fungal plant pathogens causing disease on host plants.

ORNL scientists will present new technologies available for licensing during the annual Technology Innovation Showcase. The event is 9 a.m. to 3 p.m. Thursday, June 16, at the Manufacturing Demonstration Facility at ORNL’s Hardin Valley campus.
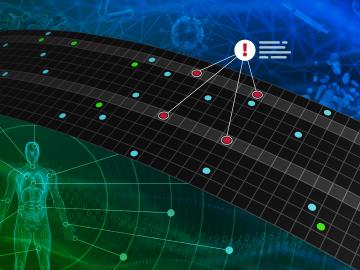
A team of researchers has developed a novel, machine learning–based technique to explore and identify relationships among medical concepts using electronic health record data across multiple healthcare providers.
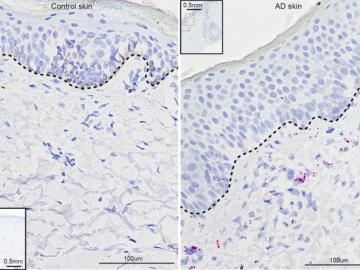
University of Pennsylvania researchers called on computational systems biology expertise at Oak Ridge National Laboratory to analyze large datasets of single-cell RNA sequencing from skin samples afflicted with atopic dermatitis.
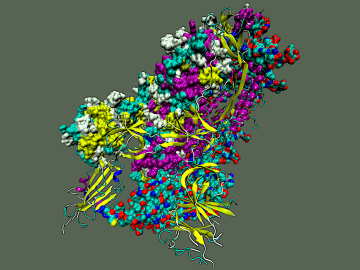
To explore the inner workings of severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2, researchers from ORNL developed a novel technique.
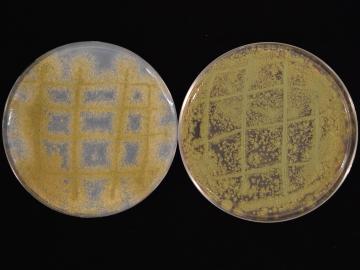
Scientists at ORNL and the University of Wisconsin–Madison have discovered that genetically distinct populations within the same species of fungi can produce unique mixes of secondary metabolites, which are organic compounds with applications in