Filter News
Area of Research
- Advanced Manufacturing (5)
- Biological Systems (1)
- Biology and Environment (46)
- Building Technologies (2)
- Clean Energy (74)
- Computational Biology (2)
- Computational Engineering (1)
- Computer Science (2)
- Electricity and Smart Grid (1)
- Energy Sciences (1)
- Functional Materials for Energy (1)
- Fusion and Fission (5)
- Fusion Energy (2)
- Isotopes (25)
- Materials (29)
- Materials for Computing (7)
- National Security (5)
- Neutron Science (13)
- Nuclear Science and Technology (6)
- Quantum information Science (1)
- Supercomputing (27)
News Topics
- (-) Biomedical (58)
- (-) Isotopes (49)
- (-) Sustainable Energy (122)
- 3-D Printing/Advanced Manufacturing (117)
- Advanced Reactors (34)
- Artificial Intelligence (88)
- Big Data (50)
- Bioenergy (88)
- Biology (96)
- Biotechnology (21)
- Buildings (55)
- Chemical Sciences (60)
- Clean Water (29)
- Climate Change (95)
- Composites (25)
- Computer Science (184)
- Coronavirus (46)
- Critical Materials (25)
- Cybersecurity (35)
- Decarbonization (75)
- Education (4)
- Element Discovery (1)
- Emergency (2)
- Energy Storage (108)
- Environment (192)
- Exascale Computing (36)
- Fossil Energy (5)
- Frontier (41)
- Fusion (53)
- Grid (61)
- High-Performance Computing (83)
- Hydropower (11)
- Irradiation (3)
- ITER (7)
- Machine Learning (46)
- Materials (141)
- Materials Science (137)
- Mathematics (6)
- Mercury (12)
- Microelectronics (2)
- Microscopy (51)
- Molten Salt (8)
- Nanotechnology (60)
- National Security (60)
- Net Zero (12)
- Neutron Science (130)
- Nuclear Energy (105)
- Partnerships (41)
- Physics (59)
- Polymers (31)
- Quantum Computing (31)
- Quantum Science (66)
- Renewable Energy (2)
- Security (24)
- Simulation (45)
- Software (1)
- Space Exploration (25)
- Statistics (3)
- Summit (57)
- Transformational Challenge Reactor (7)
- Transportation (94)
Media Contacts
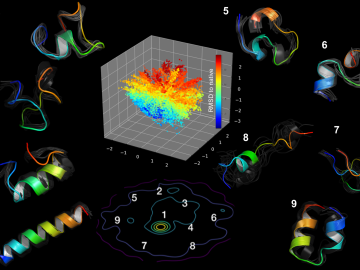
Using artificial neural networks designed to emulate the inner workings of the human brain, deep-learning algorithms deftly peruse and analyze large quantities of data. Applying this technique to science problems can help unearth historically elusive solutions.
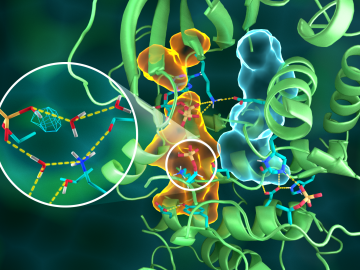
OAK RIDGE, Tenn., March 20, 2019—Direct observations of the structure and catalytic mechanism of a prototypical kinase enzyme—protein kinase A or PKA—will provide researchers and drug developers with significantly enhanced abilities to understand and treat fatal diseases and neurological disorders such as cancer, diabetes, and cystic fibrosis.
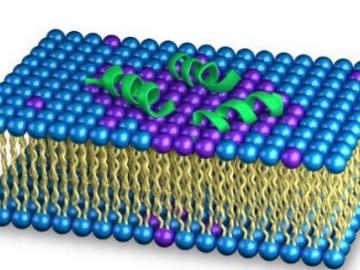
As the rise of antibiotic-resistant bacteria known as superbugs threatens public health, Oak Ridge National Laboratory’s Shuo Qian and Veerendra Sharma from the Bhaba Atomic Research Centre in India are using neutron scattering to study how an antibacterial peptide interacts with and fights harmful bacteria.

OAK RIDGE, Tenn., March 4, 2019—A team of researchers from the Department of Energy’s Oak Ridge National Laboratory Health Data Sciences Institute have harnessed the power of artificial intelligence to better match cancer patients with clinical trials.
![Coexpression_hi-res_image[1].jpg Coexpression_hi-res_image[1].jpg](/sites/default/files/styles/list_page_thumbnail/public/Coexpression_hi-res_image%5B1%5D_0.jpg?itok=OnLe-krT)
While studying the genes in poplar trees that control callus formation, scientists at Oak Ridge National Laboratory have uncovered genetic networks at the root of tumor formation in several human cancers.

OAK RIDGE, Tenn., Jan. 31, 2019—A new electron microscopy technique that detects the subtle changes in the weight of proteins at the nanoscale—while keeping the sample intact—could open a new pathway for deeper, more comprehensive studies of the basic building blocks of life.
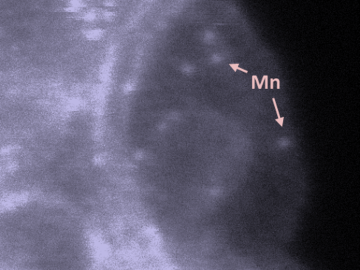
Oak Ridge National Laboratory scientists studying fuel cells as a potential alternative to internal combustion engines used sophisticated electron microscopy to investigate the benefits of replacing high-cost platinum with a lower cost, carbon-nitrogen-manganese-based catalyst.
![2018-P07635 BL-6 user - Univ of Guelph-6004R_sm[2].jpg 2018-P07635 BL-6 user - Univ of Guelph-6004R_sm[2].jpg](/sites/default/files/styles/list_page_thumbnail/public/2018-P07635%20BL-6%20user%20-%20Univ%20of%20Guelph-6004R_sm%5B2%5D.jpg?itok=DUdZNt_q)
A team of scientists, led by University of Guelph professor John Dutcher, are using neutrons at ORNL’s Spallation Neutron Source to unlock the secrets of natural nanoparticles that could be used to improve medicines.
Physicists turned to the “doubly magic” tin isotope Sn-132, colliding it with a target at Oak Ridge National Laboratory to assess its properties as it lost a neutron to become Sn-131.
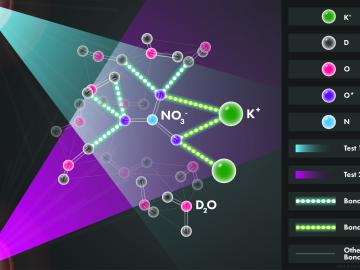
Scientists at the Department of Energy’s Oak Ridge National Laboratory used neutrons, isotopes and simulations to “see” the atomic structure of a saturated solution and found evidence supporting one of two competing hypotheses about how ions come




