
Filter News
Area of Research
News Topics
- (-) Biotechnology (24)
- (-) Molten Salt (9)
- (-) National Security (72)
- (-) Quantum Computing (37)
- 3-D Printing/Advanced Manufacturing (128)
- Advanced Reactors (34)
- Artificial Intelligence (101)
- Big Data (60)
- Bioenergy (92)
- Biology (101)
- Biomedical (61)
- Buildings (67)
- Chemical Sciences (73)
- Clean Water (31)
- Climate Change (105)
- Composites (30)
- Computer Science (198)
- Coronavirus (46)
- Critical Materials (29)
- Cybersecurity (35)
- Decarbonization (85)
- Education (5)
- Element Discovery (1)
- Emergency (2)
- Energy Storage (112)
- Environment (200)
- Exascale Computing (42)
- Fossil Energy (6)
- Frontier (45)
- Fusion (58)
- Grid (66)
- High-Performance Computing (93)
- Hydropower (11)
- Irradiation (3)
- Isotopes (57)
- ITER (7)
- Machine Learning (50)
- Materials (147)
- Materials Science (146)
- Mathematics (9)
- Mercury (12)
- Microelectronics (4)
- Microscopy (51)
- Nanotechnology (60)
- Net Zero (14)
- Neutron Science (137)
- Nuclear Energy (111)
- Partnerships (51)
- Physics (64)
- Polymers (33)
- Quantum Science (72)
- Renewable Energy (2)
- Security (25)
- Simulation (51)
- Software (1)
- Space Exploration (25)
- Statistics (3)
- Summit (59)
- Sustainable Energy (130)
- Transformational Challenge Reactor (7)
- Transportation (99)
Media Contacts

In front of family and friends, Lt. Col. Jessica Critcher and Maj. Micah McCracken gave their final report on their eye-opening year as ORNL military fellows.

ORNL scientists will present new technologies available for licensing during the annual Technology Innovation Showcase. The event is 9 a.m. to 3 p.m. Thursday, June 16, at the Manufacturing Demonstration Facility at ORNL’s Hardin Valley campus.
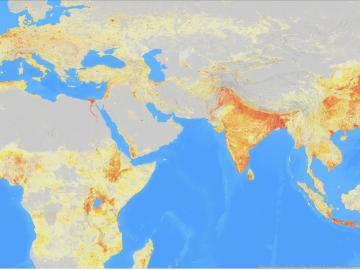
It’s a simple premise: To truly improve the health, safety, and security of human beings, you must first understand where those individuals are.

ORNL researchers used the nation’s fastest supercomputer to map the molecular vibrations of an important but little-studied uranium compound produced during the nuclear fuel cycle for results that could lead to a cleaner, safer world.
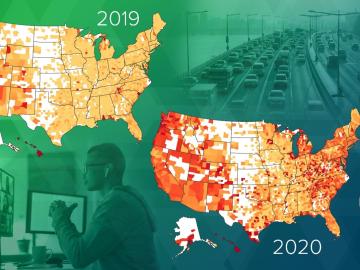
Researchers at Oak Ridge National Laboratory have empirically quantified the shifts in routine daytime activities, such as getting a morning coffee or takeaway dinner, following safer at home orders during the early days of the COVID-19 pandemic.

An Oak Ridge National Laboratory team developed a novel technique using sensors to monitor seismic and acoustic activity and machine learning to differentiate operational activities at facilities from “noise” in the recorded data.

Oak Ridge National Laboratory is debuting a small satellite ground station that uses high-performance computing to support automated detection of changes to Earth’s landscape.
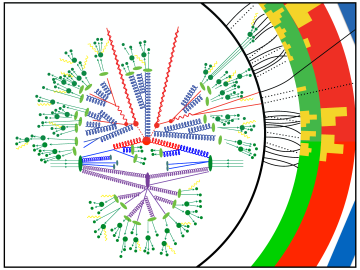
Lawrence Berkeley National Laboratory physicists Christian Bauer, Marat Freytsis and Benjamin Nachman have leveraged an IBM Q quantum computer through the Oak Ridge Leadership Computing Facility’s Quantum Computing User Program to capture part of a
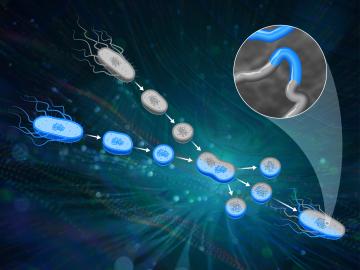
ORNL scientists had a problem mapping the genomes of bacteria to better understand the origins of their physical traits and improve their function for bioenergy production.
Scientists’ increasing mastery of quantum mechanics is heralding a new age of innovation. Technologies that harness the power of nature’s most minute scale show enormous potential across the scientific spectrum


