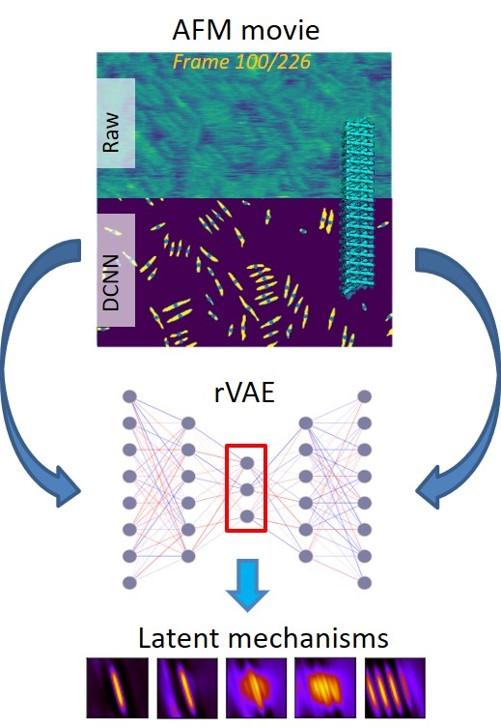
Schematic of the developed workflow.
Scientific Achievement
A machine learning workflow was developed for studying the dynamics of complex ordering systems with active rotational degrees of freedom.
Significance and Impact
The developed workflow is universally applicable for the description of optical, scanning probe, and electron microscopy imaging data and seeks to understand the dynamics of complex systems where rotations are a significant part of the process.




