Accomplishment: GCNN models were trained on two large-scale open-source datasets [2,3] that describes the HOMO-LUMO gap, which is the energy difference between the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), for millions of molecules as a function of the molecular structure. The GCNN performs a rapid and inexpensive screening of molecular structures and chemical compositions in large chemical search spaces by exploiting graphical structures of molecules defined by 1) the number of atoms in the molecule, 2) the connectivity of atoms through interatomic bonds, and 3) their chemical composition. The inexpensive but still accurate GCNN predictions of the HOMO-LUMO gap allow performing numerical optimization for molecular design that overcomes the computational limitations of existing quantum chemistry simulations to effectively identify molecules with improved photo-optical properties which would eventually open the path for more complex drug discoveries.
Deep learning (DL) models require large volumes of data for training to attain high accuracy and thus benefit an effective screening of molecules in a high dimensional space. Currently, ORNL scientists from the Computational Sciences and Engineering Division (CSED) are leveraging the CADES cluster and OLCF supercomputing resources to run density functional theory tight-binding (DFTB) simulations [1] to collect quantum chemistry data about molecules that thoroughly span high dimensional molecular spaces.
On such large volumes of data, DL models cannot be trained in sequential mode, as it would require several days/weeks for the training to be completed. Therefore, high-performance DL training that can effectively utilize current and next generation supercomputing resources is needed to accelerate the learning process.
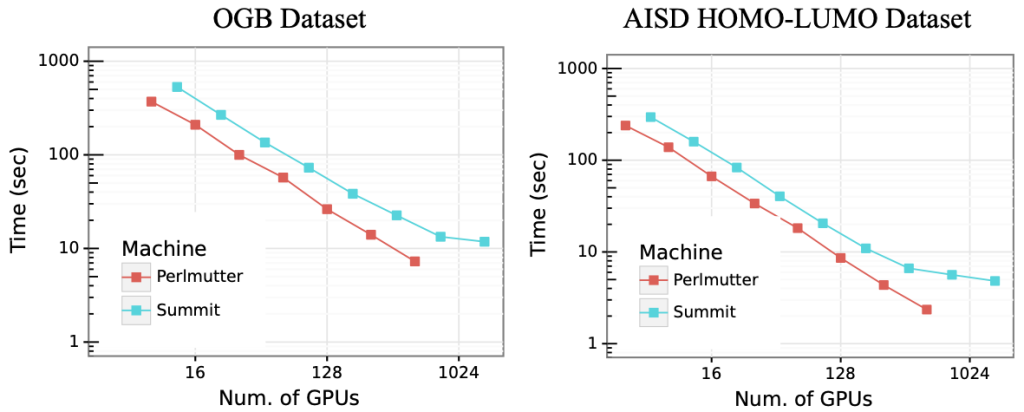
To develop a high-performance deep learning workflow, we use HydraGNN [4], an open source distributed implementation [5] of multi-headed graph convolutional neural networks, along with ADIOS, a high-performance data management framework for large-scale data. We use ADIOS to scale the reading of molecular data from millions of files, and use HydraGNN’s distributed data parallelism capability to scale the training of the model on hybrid CPU/GPU multi-node architectures. We train GCNN models on two open-source molecular graph datasets: the Open Graph Benchmark (OGB) dataset [2] and the AISD HOMO-LUMO dataset [3], consisting of 3 and 11 million graphs, respectively. We test the scalability of I/O and GCNN training on Summit and Perlmutter. Numerical results show that the training of HydraGNN scales linearly up to 1,024 GPUs on both systems.
Acknowledgement: This research is funded by the AI Initiative, as part of the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the U.S. Department of Energy (DOE).
Contact: Massimiliano (Max) Lupo Pasini (lupopasinim@ornl.gov)
Team: Jong Youl Choi, Pei Zhang, Kshitij Mehta, Andrew Blanchard, Massimiliano (Max) Lupo Pasini
References:
- Gaus, M., Goez, A., Elstner, M.: Parametrization and benchmark of DFTB3 for organic molecules. Journal of Chemical Theory and Computation 9(1), 338–354 (2013). https://doi.org/10.1021/ct300849w
- Hu, W., Fey, M., Zitnik, M., Dong, Y., Ren, H., Liu, B., Catasta,M., Leskovec, J.: Open graph benchmark: Datasets for machine learning on graphs. Advances in Neural Information Processing Systems 2020-December (NeurIPS), 1–34 (2020)
- Blanchard, A.E., Gounley, J., Bhowmik, D., Pilsun, Y., Irle, S.: AISD HOMO-LUMO. https://doi.org/10.13139/ORNLNCCS/1869409
- Lupo Pasini, M., Zhang, P., Reeve, S.T., Choi, J.Y.: Multi-task graph neural networks for simultaneous prediction of global and atomic properties in ferromagnetic systems. Machine Learning: Science and Technology. 3(2), 025007(2022). https://doi.org/10.1088/2632-2153/ac6a51
- Lupo Pasini, M., Reeve, S.T., Zhang, P., Choi, J.Y.: HydraGNN. [Computer Software] https://doi.org/10.11578/dc.20211019.2 (2021) https://github.com/ORNL/HydraGNN






